Our approach to drug discovery is engineered to identify best-in-class small molecule therapeutics for disease areas where effective therapeutic solutions are lacking.
Combining Physics, Chemistry, Biology and Computation
Our proprietary QUAISAR platform enables us to design drug candidates by combining computational physics and artificial intelligence with expertise in disease biology, chemistry, biophysics, proteomics, and translational informatics. We focus on protein targets with strong biological and genetic validation in order to design molecules that rapidly reach the desired target product profile (TPP)
We perform biophysical and structural biology studies in our laboratory to characterize the most promising protein-ligand complexes. As data accumulate on our projects, we integrate machine learning to improve the accuracy and applicability of the property prediction models. Our integrated multi-disciplinary drug discovery team focuses on the key properties of our molecules to systematically engineer, in an atom-by-atom fashion, lead candidates that achieve the desired TPP, overcoming limitations in existing chemical matter and, ultimately, delivering best-in-class medicines to patients. It is the predictive power of QUAISAR, coupled with our multi-disciplinary team, that presents the opportunity to propel our projects rapidly toward the clinic.
Validated Approach
We have validated our QUAISAR-driven computation-first approach via Stimulator of Interferon Genes (STING), where we uniquely solved the problem of designing a small molecule with drug-like properties for a large, charged, undrugged binding site. SNX281 is currently in clinical development at a spinout company for patients with advanced solid tumors and lymphoma.
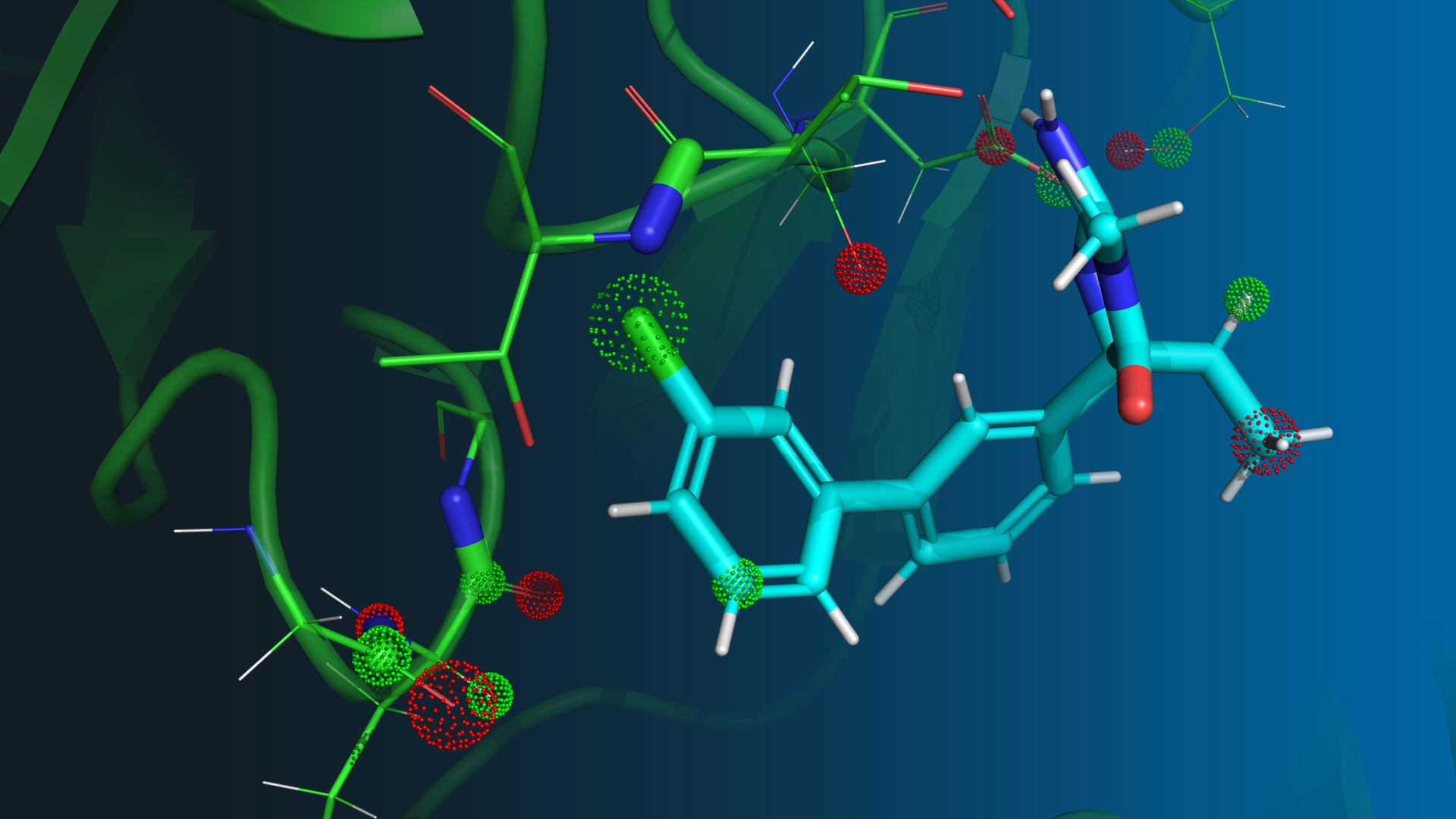
Accurate All-Atom Physics-Based Simulations
Traditional computational approaches to drug design treat proteins as rigid, fixed molecules. But, in living organisms, proteins are shape-shifting and highly dynamic. Understanding this motion and accurately predicting the relevant thermodynamic properties enables us to engineer molecules with desirable properties, atom-by-atom. We perform hundreds of quantum mechanical calculations on each ligand to measure required parameters and develop a highly accurate molecular force field. We use this force field to run long-timescale molecular dynamics simulations of biologically relevant systems including proteins, ligands, cofactors, membranes and ions.
Machine Learning Models
We build highly accurate property prediction models based on curated literature data. As data accumulate over the course of a project, we augment our models with a bias toward the local chemical space of interest. We also leverage machine learning to rapidly explore chemical space through generative models and reaction-based enumerations, which are filtered through our physics-based computational assays to select most promising molecules for synthesis.
Integrated Team, Rapid Iterations
We minimize the design cycle time by incorporating knowledge about the difficulty of chemical synthesis and availability of chemical building blocks. We perform simulations on massive internal high-performance computing (HPC) resources to deliver decision-making results in a timely fashion. Our nimble project teams with deep domain expertise work in unison to make data-driven decisions that drive our projects toward the desired TPP.